Orange: Linear Projection: Difference between revisions
Onnowpurbo (talk | contribs) Created page with "Sumber: https://docs.biolab.si//3/visual-programming/widgets/visualize/linearprojection.html A linear projection method with explorative data analysis. Inputs Data: i..." |
Onnowpurbo (talk | contribs) No edit summary |
||
| Line 8: | Line 8: | ||
Data: input dataset | Data: input dataset | ||
Data Subset: subset of instances | Data Subset: subset of instances | ||
Projection: custom projection vectors | Projection: custom projection vectors | ||
| Line 16: | Line 14: | ||
Selected Data: instances selected from the plot | Selected Data: instances selected from the plot | ||
Data: data with an additional column showing whether a point is selected | Data: data with an additional column showing whether a point is selected | ||
Components: projection vectors | Components: projection vectors | ||
| Line 25: | Line 21: | ||
Consider, for a start, a projection of the Iris dataset shown below. Notice that it is the sepal width and sepal length that already separate Iris setosa from the other two, while the petal length is the attribute best separating Iris versicolor from Iris virginica. | Consider, for a start, a projection of the Iris dataset shown below. Notice that it is the sepal width and sepal length that already separate Iris setosa from the other two, while the petal length is the attribute best separating Iris versicolor from Iris virginica. | ||
[[File:Linear-projection-stamped.png|center|200px|thumb]] | |||
Axes in the projection that are displayed and other available axes. | Axes in the projection that are displayed and other available axes. | ||
Optimize your projection by using Suggest Features. This feature scores attributes by average classification accuracy and returns the top scoring attributes with a simultaneous visualization update. | Optimize your projection by using Suggest Features. This feature scores attributes by average classification accuracy and returns the top scoring attributes with a simultaneous visualization update. | ||
Choose the type of projection. | Choose the type of projection. | ||
Axes inside a circle are hidden. Circle radius can be be changed using a slider. | Axes inside a circle are hidden. Circle radius can be be changed using a slider. | ||
Adjust plot properties: | Adjust plot properties: | ||
Set jittering to prevent the dots from overlapping (especially for discrete attributes). | Set jittering to prevent the dots from overlapping (especially for discrete attributes). | ||
Show legend displays a legend on the right. Click and drag the legend to move it. | Show legend displays a legend on the right. Click and drag the legend to move it. | ||
Show class density colors the graph by class (see the screenshot below). | Show class density colors the graph by class (see the screenshot below). | ||
Label only selected points allows you to select individual data instances and label them. | Label only selected points allows you to select individual data instances and label them. | ||
Select, zoom, pan and zoom to fit are the options for exploring the graph. Manual selection of data instances works as an angular/square selection tool. Double click to move the projection. Scroll in or out for zoom. | Select, zoom, pan and zoom to fit are the options for exploring the graph. Manual selection of data instances works as an angular/square selection tool. Double click to move the projection. Scroll in or out for zoom. | ||
If Send automatically is ticked, changes are communicated automatically. Alternatively, press Send. | If Send automatically is ticked, changes are communicated automatically. Alternatively, press Send. | ||
Save Image saves the created image to your computer in a .svg or .png format. Produce a report. | Save Image saves the created image to your computer in a .svg or .png format. Produce a report. | ||
==Contoh== | |||
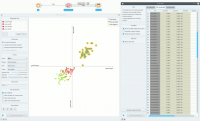
The Linear Projection widget works just like other visualization widgets. Below, we connected it to the File widget to see the set projected on a 2-D plane. Then we selected the data for further analysis and connected it to the Data Table widget to see the details of the selected subset. | The Linear Projection widget works just like other visualization widgets. Below, we connected it to the File widget to see the set projected on a 2-D plane. Then we selected the data for further analysis and connected it to the Data Table widget to see the details of the selected subset. | ||
[[File:LinearProjection-example.png|center|200px|thumb]] | |||
==Referensi== | |||
Koren Y., Carmel L. (2003). Visualization of labeled data using linear transformations. In Proceedings of IEEE Information Visualization 2003, (InfoVis’03). Available here. | Koren Y., Carmel L. (2003). Visualization of labeled data using linear transformations. In Proceedings of IEEE Information Visualization 2003, (InfoVis’03). Available here. | ||
Revision as of 07:42, 22 January 2020
Sumber: https://docs.biolab.si//3/visual-programming/widgets/visualize/linearprojection.html
A linear projection method with explorative data analysis.
Inputs
Data: input dataset Data Subset: subset of instances Projection: custom projection vectors
Outputs
Selected Data: instances selected from the plot Data: data with an additional column showing whether a point is selected Components: projection vectors
This widget displays linear projections of class-labeled data. It supports various types of projections such as circular, linear discriminant analysis, principal component analysis, and custom projection.
Consider, for a start, a projection of the Iris dataset shown below. Notice that it is the sepal width and sepal length that already separate Iris setosa from the other two, while the petal length is the attribute best separating Iris versicolor from Iris virginica.

Axes in the projection that are displayed and other available axes.
Optimize your projection by using Suggest Features. This feature scores attributes by average classification accuracy and returns the top scoring attributes with a simultaneous visualization update.
Choose the type of projection.
Axes inside a circle are hidden. Circle radius can be be changed using a slider.
Adjust plot properties:
Set jittering to prevent the dots from overlapping (especially for discrete attributes).
Show legend displays a legend on the right. Click and drag the legend to move it.
Show class density colors the graph by class (see the screenshot below).
Label only selected points allows you to select individual data instances and label them.
Select, zoom, pan and zoom to fit are the options for exploring the graph. Manual selection of data instances works as an angular/square selection tool. Double click to move the projection. Scroll in or out for zoom.
If Send automatically is ticked, changes are communicated automatically. Alternatively, press Send.
Save Image saves the created image to your computer in a .svg or .png format. Produce a report.
Contoh
The Linear Projection widget works just like other visualization widgets. Below, we connected it to the File widget to see the set projected on a 2-D plane. Then we selected the data for further analysis and connected it to the Data Table widget to see the details of the selected subset.
Referensi
Koren Y., Carmel L. (2003). Visualization of labeled data using linear transformations. In Proceedings of IEEE Information Visualization 2003, (InfoVis’03). Available here.
Boulesteix A.-L., Strimmer K. (2006). Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Briefings in Bioinformatics, 8(1), 32-44. Abstract here.